Introduction
OrganSegC2F (Organ Segmentation Coarse-to-Fine) is aimed at efficient organ segmentation from CT scan.
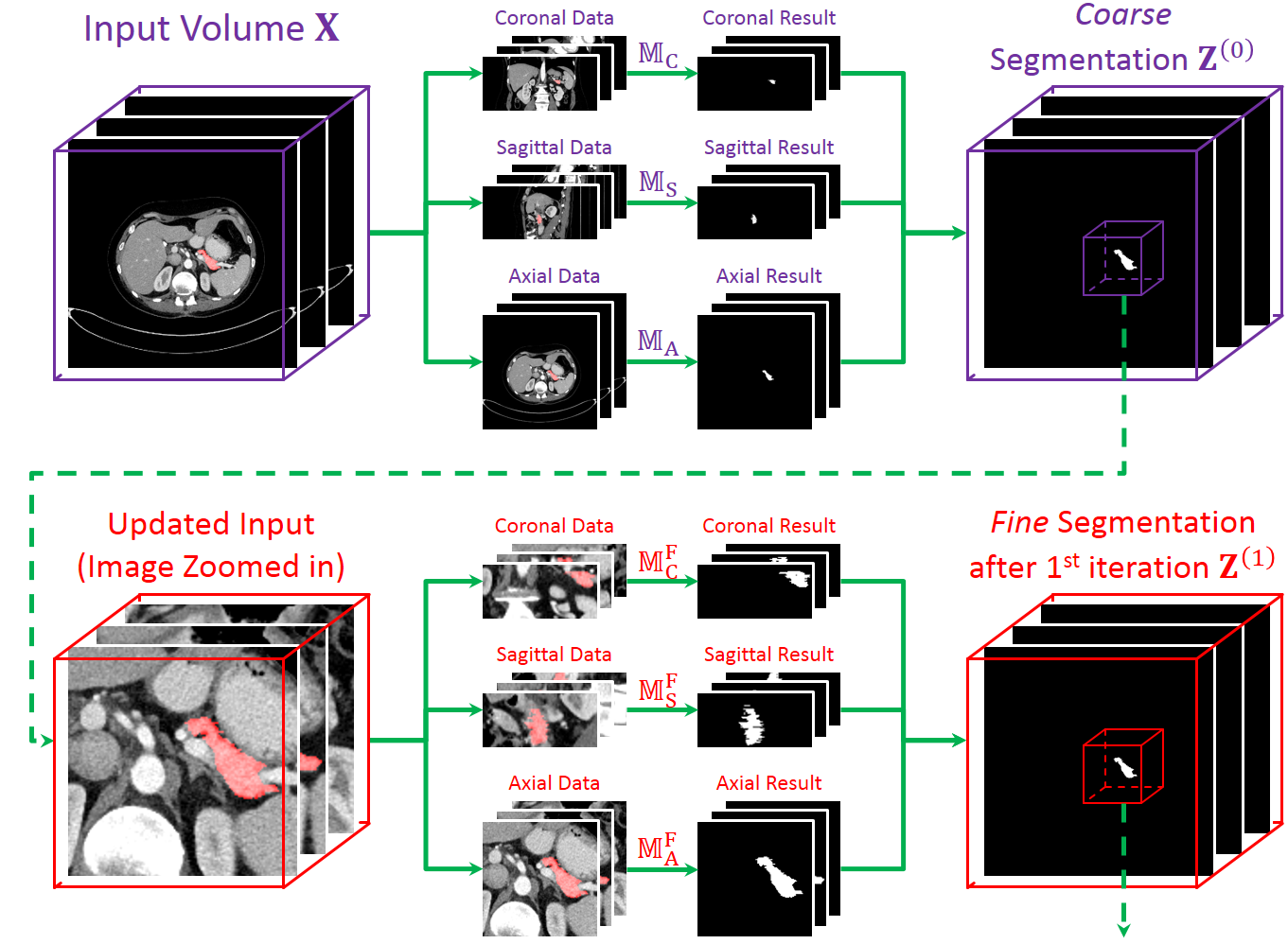
It is based on a 2D deep segmentation network (currently FCN [Long, CVPR15] is used).
The 3D abdominal CT scan is sliced into 2D pieces, fed into the network, and the results are stacked back to a 3D volume.
Our approach is motivated by the fact that some organs, e.g., the pancreas, are quite small in volume,
and they often occupy a small region in each slice of CT scans.
Using these data for training and testing may cause a deep network disrupted by the background noise.
Thus, we train two set of networks, one for coarse-scaled and one for fine-scaled segmentation.
In the testing process, we use the coarse-scaled networks to generate an initial segmentation,
and apply the fine-scaled networks to fine-tuning segmentation through several iterations.
Our approach works very well on the NIH pancreas segmentation dataset [Roth, MICCAI15].
We outperform the state-of-the-art [Roth, MICCAI16] by more than 4%.
Moreover, the fine-tuning stage helps to improve the accuracy of the worst case,
with an impressive accuracy gain of over 20%.
The accuracy on the worst case is over 60%, which guarantees the reliability in clinical applications.
Frequently Asked Questions
- What kind of data does OrganSegC2F work on?
OrganSegC2F was designed for abdominal CT scan,
but theoretically, it can also work for other types of 3D data, like fMRI.
- How long does it take to process one volume at the testing stage?
It depends on the volume size and the graphical card you use (we use one Titan-X Maxwell).
In the NIH dataset, an average volume size is around 512x512x300, and we need an average of 3 minutes.
The time grows almost linearly as the number of voxels in the volume.
- If I want to use the codes for my experiments, what paper shall I cite?
Please cite our MICCAI paper.
Downloads
- For detailed guidance for running the codes, please refer to the README file in each code package.
- The codes are to be used with a CAFFE library.
Please find the Dice loss layer in the dice_loss_layer.{hpp/cpp} files and put them in CAFFE
(currently no GPU implementation is available but it is pretty cheap in computation).
- CURRENT VERSION!
[RAR package][ZIP package]
Version 1.10, released on October 19, 2017.
View this change log.
- OLD VERSIONS.
[RAR package][ZIP package]
Version 1.9, released on October 1, 2017.
[RAR package][ZIP package]
Version 1.8, released on September 22, 2017.
[RAR package][ZIP package]
Version 1.7, released on September 21, 2017.
[RAR package][ZIP package]
Version 1.6, released on July 11, 2017.
[RAR package][ZIP package]
Version 1.5, released on July 06, 2017.
[RAR package][ZIP package]
Version 1.4, released on June 29, 2017.
[RAR package][ZIP package]
Version 1.3, released on June 27, 2017.
[RAR package][ZIP package]
Version 1.2, released on June 18, 2017.
[RAR package][ZIP package]
Version 1.1, released on June 11, 2017.
[RAR package][ZIP package]
Version 1.0, released on June 01, 2017.
- If you find any mistakes or bugs in the codes or datasets, please contact me.
Instructions to Reproduce the Results in Our Paper
In the README file, you can see a detailed, step-by-step guideline.
- Download the NIH pancreas segmentation dataset from
this website
(please cite their paper [Roth, MICCAI15] accordingly).
- Make sure that you are equipped with a modern GPU and it supports CAFFE.
- Download our library, and make the CAFFE and its python interface.
- Transfer the data into numpy (NPY) files using our tool (released in the library) or your own tool.
You should have 82 image files and 82 label files, stored in the directory specified by the program.
- In run.sh, specify the global variables especially those related to path.
- Execute each module orderly.
Related Publications
- Please cite our MICCAI paper rather than the earlier version put on arXiv. Thanks!
- Yuyin Zhou, Lingxi Xie, Wei Shen, Yan Wang, Elliot Fishman and Alan Yuille,
"A Fixed-Point Model for Pancreas Segmentation in Abdominal CT Scans",
in International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI),
Quebec City, Quebec, Canada, 2017.
[PDF]
[BibTeX]
- Yuyin Zhou, Lingxi Xie, Wei Shen, Elliot Fishman and Alan Yuille,
"Pancreas Segmentation in Abdominal CT Scan: A Coarse-to-Fine Approach",
arXiv preprint arXiv: 1612.08230.
References
- [Roth, MICCAI16] Holger Roth, Le Lu, Amal Farag, Andrew Sohn, and Ronald M. Summers,
"Spatial Aggregation of Holistically-Nested Networks for Automated Pancreas Segmentation",
in International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), 2016.
- [Roth, MICCAI15] Holger Roth, Le Lu, Amal Farag, Hoo-chang Shin, Jiamin Liu, Evrim Turkbey, and Ronald Summers,
"DeepOrgan: Multi-level Deep Convolutional Networks for Automated Pancreas Segmentation",
in International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), 2015.
- [Long, CVPR15] Jonathan Long, Evan Shelhamer, and Trevor Darrell,
"Fully Convolutional Networks for Semantic Segmentation",
in IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2015.